Many cells in the human body migrate to perform their function. Immune cells move through the body to fight infections and control intruders. Cancer cells move and spread cancer. Stem cells move to damaged tissue, differentiate and build new tissue. Understanding cell migration is fundamentally important and essential for identifying targets for novel therapies.
Cancer cells and stem cells are known to "choose" the more efficient of two migration modes: mesenchymal or amoeboid. Experiments in 2D cell cultures have shown that the mesenchymal amoeboid transition (MAT) can be triggered by physical stimuli and that different physical and chemical processes in the cytoskeleton dominate each mode.
The goals of this project are 1) to establish which features of a 3D extracellular matrix (ECM) trigger MAT and 2) if the transition can be modelled as a physicochemical instability in the cytoskeleton (as opposed to changes in gene expression). This will be studied by combining state of the art in vitro cell imaging experiments and mathematical modelling:
- microfluidic devices with embedded biomimetic hydrogel (3D ECM) to accurately control fluids and ECM deformation while allowing high resolution imaging in 5D(XYZ, lambda and t).
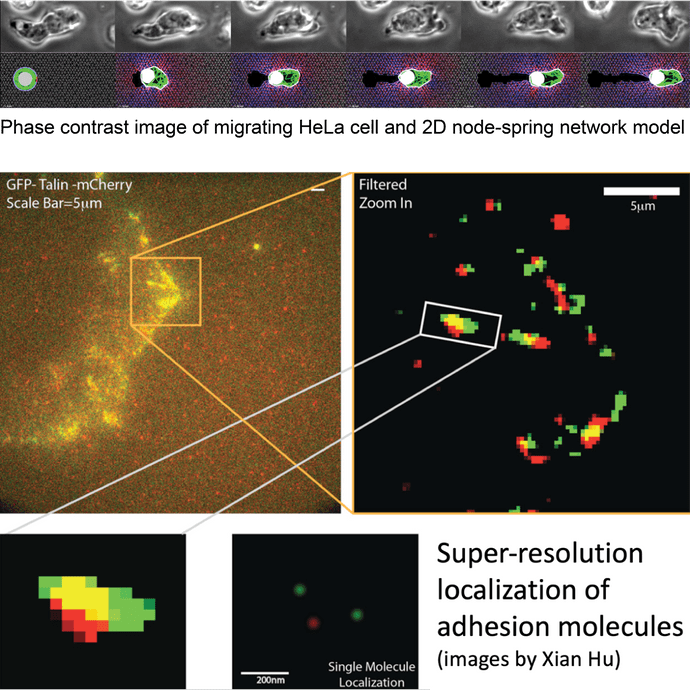
- state of the art fluorescence live cell super-resolution microscopy (Airyscan, iSIM and SPIM), phase contrast microscopy and quantitative image analysis.
- a 3D agent-based model using node-spring networks to represent both the 3D ECM, actin-myosin cytoskeleton network and the cell membrane.
- parameters of the ECM and cell contractility and adhesion will be co-varied in model and experiments to tune the model.
Requirements
- The candidate must be motivated to develop both experimental and numerical tools for the project and have a MSc in physics, mathematics or computer science or a MSc in life science with experience in mathematical modelling or scientific programming.
- Candidates with documented experience in scientific programming, machine learning, live cell imaging and microfluidics will be prioritized.
Supervisors
Professor Anders Malthe-Sørenssen
Call 2: Project start autumn 2022
This project is in call 2, starting autumn 2022. Read about how to apply