Reliable determination of diagnostic proteins (i.e. biomarkers) and biopharmaceuticals in serum, plasma or blood is a challenging task. The composition of the sample is very complex and protein vary in their concentration level from mg/mL level (like albumin, immunoglobulins) down to femtogram/mL level (like interleukins).
Often the concentration of the biomarkers we want to measure is extremely low and the difficulties arise when we want to determine these valuable markers in the presence of so-called high-abundant proteins like albumin (mg/mL level).
We have been working on several concepts to allow reliable determination of these ultra-low-abundant proteins: immunocapture of biomarker proteins using monoclonal antibodies, fishing of epitope-containing peptides using monoclonal antibodies and sample clean-up and enrichment using “plastic” antibodies (so-called molecularly imprinted polymers (MIP)).
Immunocapture using monoclonal antibodies
This is the concept we have been working on for the longest time. The figure below shows how this strategy is applied.
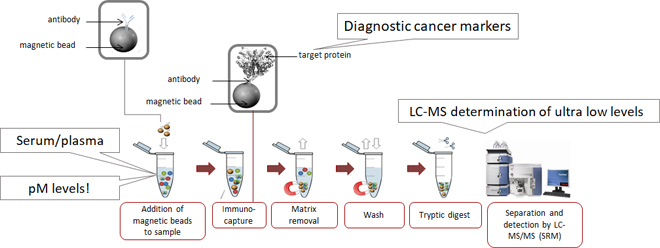
Simply explained, the biomarker of interest is very selectively captured by monoclonal antibodies which are attached to small paramagnetic beads (2.8 µm in size, invisibly small for the human eye). After our biomarker has bound to the antibody, it is literally pulled out of the complex biological sample using a magnet. In this way we get rid of (almost) all the interfering proteins. After a descent wash, we treat the proteins (our biomarker) still attached to the beads with trypsin. This treatment cuts our biomarker very reproducible into small but highly specific peptides. These specific peptides are then measured by state of the art LC-MS.
In this way we have been able to successfully determine several biomarkers for lung cancer, ovarium cancer and testicular cancer at clinical relevant levels.
Fishing of epitope-containing peptides using monoclonal antibodies
This a new concept, which we have been working on for a couple of years. Here we focus very much on the epitope of the biomarker. The epitope is the small part of the protein (6-10 amino acids) to which the monoclonal antibody binds. The procedure resembles very much immunocapture as shown in the figure above. The difference lies in the order of the steps. See the figure below for the difference in concept.

As mentioned for immunocapture, before we measure the proteins, we treat them with trypsin. This to cut (digest) the proteins in small but highly specific peptides. In the concept of fishing epitope-containing peptides, the serum, plasma or blood sample is first treated with trypsin, yielding a very complex mixture of tryptic peptides. Monoclonal antibodies bound to magnetic beads are used to capture only those peptides containing the epitope of the biomarker. The magnetic beads are then pulled out of the complex peptide mixture, and the epitope-containing peptides which remained on the antibodies after washing are analyzed using LC-MS.
Enrichment using “plastic” antibodies (also called molecularly imprinted polymers; MIPs)
Our group has been collaborating within several European networks since 2011 to study these MIPs in biomarker analysis. From fishing epitope-containing peptides using monoclonal antibodies to peptide extraction using MIPs is a small step. The only difference is that instead of monoclonal antibodies, MIPs are used to extract the small but specific peptides from a digested biological sample. Our collaborators in the European network BioCapture are able to synthesize small particles containing cavities in which only certain specific peptides can bind. This makes these MIPs an excellent research topic within our research focus.