 Atlantic cod seems to have a much higher density of STRs (short tandem repeats, also called microsatellites) than other species. STRs are sequences where a short unit of nucleotides (typically 1-10 bp) are repeated many times (typically 5-50) right after each other. For example, ACACACACAC is a dinucleotide repeat, while AGCAGCAGCAGCAGC is a trinucleotide repeat.
Atlantic cod seems to have a much higher density of STRs (short tandem repeats, also called microsatellites) than other species. STRs are sequences where a short unit of nucleotides (typically 1-10 bp) are repeated many times (typically 5-50) right after each other. For example, ACACACACAC is a dinucleotide repeat, while AGCAGCAGCAGCAGC is a trinucleotide repeat.
In the first figure (from Tørresen et al., 2017), we have compared cod (torsk, Gadus morhua, indicated with Gm) to all other species in Ensembl, a database containing genomes from many species (more than 60), with regards to STRs in the whole genome, in the promotor sequence (2 kbp upstream of genes, often involved in regulating when and how much of the gene product is made) and in the actual coding sequence (which encodes the actual protein product of the gene). Especially in the whole genome/assembly (the first panel) you see that cod is clearly separated from the other species (with up to 11% of its genome in STRs), specifically from mammals, which are marked in blue. In the last panel, you see that cod has many more STRs in coding sequences than many other species.
We have also sequenced haddock (hyse, Melanogrammus aeglefinus), and find many of the same characteristics there. In the second figure (from Tørresen et al., 2018) we compared cod and haddock to the other fishes in Ensembl.
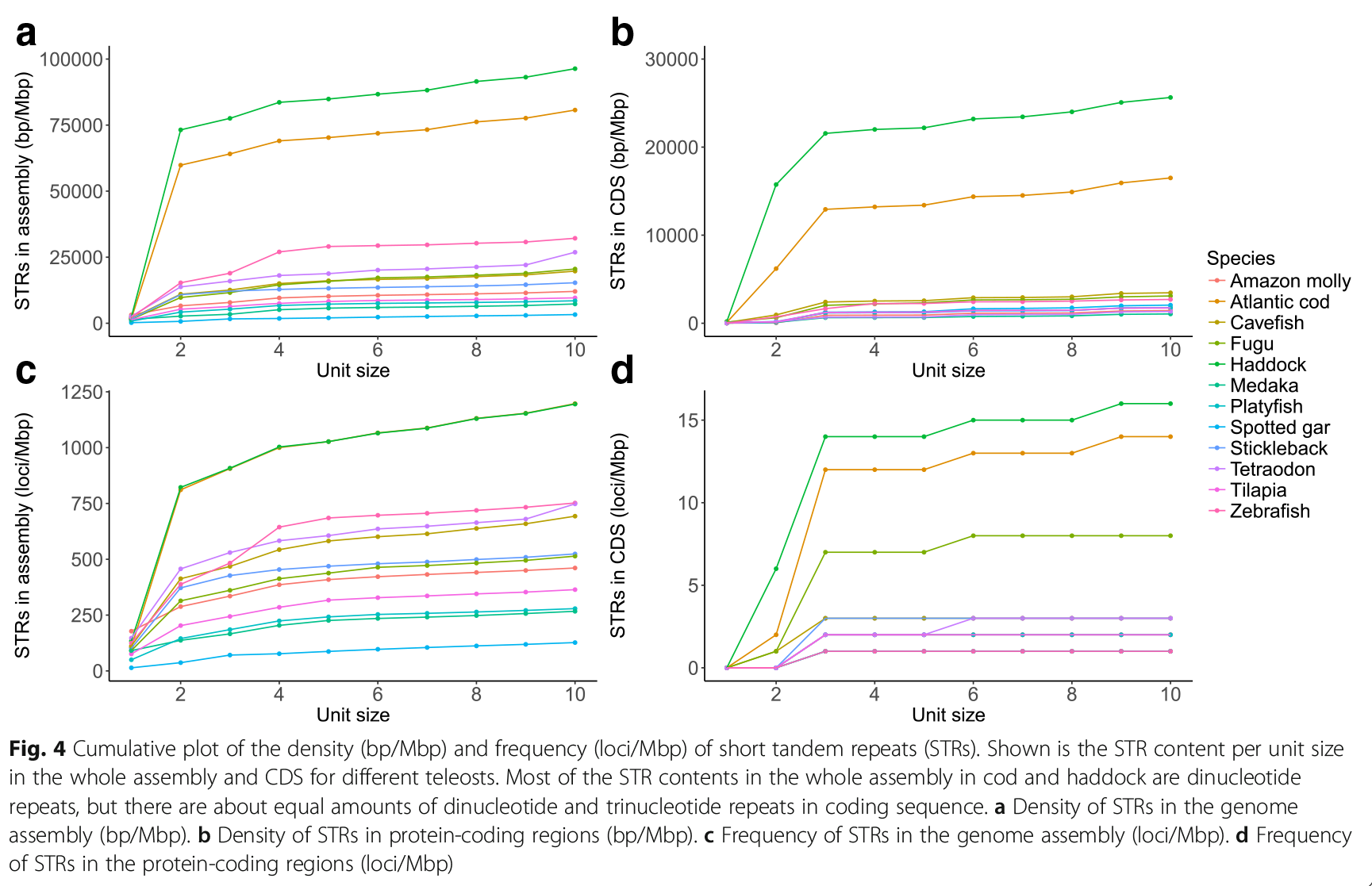
This is a cumulative plot, where the position up in the right is the sum of all the previous STRs. Then you can for instance see that most of the STRs in coding regions are in dinucleotides and trinucleotides.
Based on this, we are quite certain that codfishes, like cod and haddock, are special with regards to STRs compared to other species. We don’t exactly know why or how it has evolved. To start figuring this out, we have considered multiple approaches. One is to see which kinds of genes these STRs sit in. We have written a bit about this in the paper about the haddock genome above.
We don’t really know which features of codfishes that facilities this large amount of STRs. We speculate that the STRs can be important for rapid adaptation to the environment (they mutate at much higher frequency than regular DNA which are usually substitutions). The large effective population size of cod might also have some effect here, and possibly the method of reproduction (massive amounts of eggs in batch spawning), which means that cod can “afford” that some of the offspring do not survive because of detrimental mutations. Some of this is not trivial to find out.
The attributes mentioned above (population size, reproduction method) might help cod to tolerate (and thrive) with the large STRs burden, but they do not say anything about how STRs actually spread in the genome. In some species, like squamate reptiles, it seems that a transposable element is connected to the spread of STRs (the element itself contains STRs or STRs-like sequences that are spread when the transposable element is copied across the genome) (Pasquesi et al., 2018). A transposable element (TE) is a DNA sequence that encodes its own mobility, enabling it to spread across the genome and inserting itself in different locations. Codfishes do not have this specific transposable element (CR1-L3 LINEs), so this is not the answer to the question. However, other transposable elements may be active.
An investigation of what kinds of motifs (sequence patterns) surround STRs in codfishes would be a good start of an investigation into the spread of STRs. These motifs might connect STRs to certain TEs, or to other genomic attributes (cross-over events for instance). Also, investigation into other fishes might find motifs unique to codfishes that could help unravel this intriguing attribute.
For an overview of STRs, see Gymrek (2017). To examine these features, we will utilize strategies such as k-mer analyses (Sievers et al., 2017), where all the occurrences of any subsequence of length k in the genomes are analysed. We will identify regularities and deviations in sequence contexts. Through this and similar approaches we will map the particular features that sets the codfish apart from other species, to try to delineate the driving forces for this from a sequence context, with a perspective on evolution. Among the tools will be The Genomic HyperBrowser framework for statistical genomics (Sandve et al., 2010, Sandve et al. 2013, Simovski et al., 2017).
References
- Gymrek M (2017) A genomic view of short tandem repeats. Curr Opin Genet Dev., 44, 9-16. doi: 10.1016/j.gde.2017.01.012
- Pasquesi GIM, Adams RH, Card DC, Schield DR, Corbin AB, Perry BW, Reyes-Velasco J, Ruggiero RP, Vandewege MW, Shortt JA, Castoe TA (2018) Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun., 9(1), 2774. doi: 10.1038/s41467-018-05279-1
- Sandve GK, Gundersen S, Johansen M, Glad IK, Gunathasan K, Holden L, Holden M, Liestøl K, Nygård S, Nygaard V, Paulsen J, Rydbeck H, Trengereid K, Clancy T, Drabløs F, Ferkingstad E, Kalas M, Lien T, Rye MB, Frigessi A, Hovig E (2013) The Genomic HyperBrowser: an analysis web server for genome-scale data. Nucleic Acids Res., 41 (Web Server issue), W133-41. doi: 10.1093/nar/gkt342
- Sandve GK, Gundersen S, Rydbeck H, Glad IK, Holden L, Holden M, Liestøl K, Clancy T, Ferkingstad E, Johansen M, Nygaard V, Tøstesen E, Frigessi A, Hovig E (2010) The Genomic HyperBrowser: inferential genomics at the sequence level. Genome Biol., 11(12), R121. doi: 10.1186/gb-2010-11-12-r121
- Sievers A, Bosiek K, Bisch M, Dreessen C, Riedel J, Froß P, Hausmann M, Hildenbrand G. (2017) K-mer Content, Correlation, and Position Analysis of Genome DNA Sequences for the Identification of Function and Evolutionary Features. Genes (Basel), 8(4), e122. doi: 10.3390/genes8040122
- Simovski B, Vodák D, Gundersen S, Domanska D, Azab A, Holden L, Holden M, Grytten I, Rand K, Drabløs F, Johansen M, Mora A, Lund-Andersen C, Fromm B, Eskeland R, Gabrielsen OS, Ferkingstad E, Nakken S, Bengtsen M, Nederbragt AJ, Thorarensen HS, Akse JA, Glad I, Hovig E, Sandve GK (2017) GSuite HyperBrowser: integrative analysis of dataset collections across the genome and epigenome. Gigascience., 6(7), 1-12. doi: 10.1093/gigascience/gix032
- Tørresen OK, Brieuc MSO, Solbakken MH, Sørhus E, Nederbragt AJ, Jakobsen KS, Meier S, Edvardsen RB, Jentoft S. (2018) Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats. BMC Genomics, 19(1), 240. doi: 10.1186/s12864-018-4616-y
- Tørresen OK, Star B, Jentoft S, Reinar WB, Grove H, Miller JR, Walenz BP, Knight J, Ekholm JM, Peluso P, Edvardsen RB, Tooming-Klunderud A, Skage M, Lien S, Jakobsen KS, Nederbragt AJ (2017) An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics, 18(1), 95. doi: 10.1186/s12864-016-3448-x
Supervisors
- Eivind Hovig, Bioinformatics centre/BMI, Department of Informatics (main supervisor)
- Ole K. Tørresen, CEES, Department of Biosciences
- Kjetill S. Jakobsen, CEES, Department of Biosciences
- Torbjørn Rognes, Bioinformatics centre/BMI, Department of Informatics